Multispectral Imaging Broadens Cellular Analysis
Originating Technology/NASA Contribution
How does life begin and evolve? Does life exist elsewhere in the universe? What is the future of life on Earth and beyond?
These questions have lingered for many years, and we still do not fully know the answers to them. NASA, however, in its efforts to sustain life here on our home planet and chart a course for humans to explore the Moon, Mars, and beyond, is going to new depths to better address them—deep beneath the ocean surface.
In the floors of our oceans are holes that spew hot, gaseous, mineral-rich liquids from the deep, subsurface magma below. Scientists from NASA’s Advanced Life Support Group at Ames Research Center are hoping that these holes, called hydrothermal vents, may not only help them unlock the secrets of the deep, but help them learn if exotic life forms exist on other planetary bodies, including Mars and Jupiter’s moon Europa.
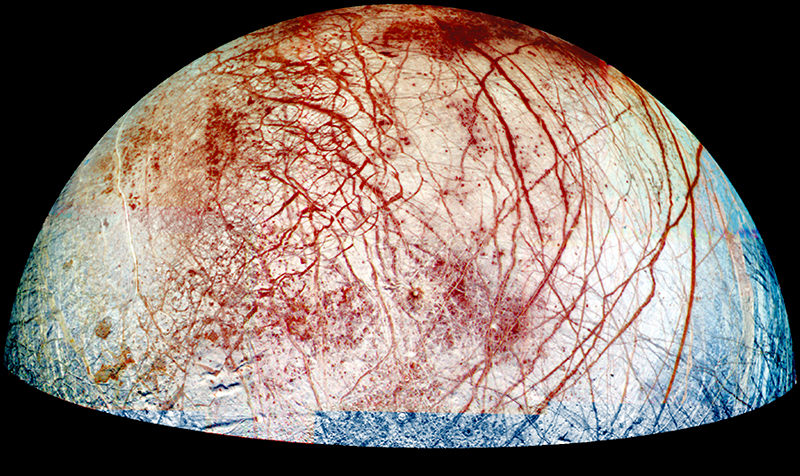
Because hydrothermal vents are thousands of meters underwater, they are not exposed to sunlight. Without sunlight, there may be abiotic life forms that exist devoid of photosynthetic input or decomposition of organic materials—in or near these vents. NASA scientists hypothesize that the deep subsurface of Earth could be home to organisms that exist solely on chemical energy that is generated from the off-gassing magma below the ocean floor. They further hope that this extreme underground environment could provide insight into whether similar life forms exist elsewhere in the universe, in environments that are far removed from the Sun and therefore also require other sources of energy. For example, scientists have targeted Europa because of its thick ice crusts and the mounting evidence that points to there being an ice-covered ocean that could potentially harbor similar hydrothermal vents.
To test their hypothesis, the Advanced Life Support Group scientists have built a life-detection instrument called Medusa to collect, store, and analyze sample organisms from erupting hydrothermal vents. For the sample analyses, Medusa is equipped with a spectral-analysis chemical sensor that uses a process called flow cytometry to examine the natural glow of light, or fluorescence, emitted from any of the samples collected as of fluorescence emitted by each cell. Because they do not have the ability to determine where in the cell the signal is coming from, their applications in cell biology are mainly limited to measuring the total quantities of specific molecules in or on the cell. A few flow cytometry systems can produce images of cells in flow using transmitted light, but these systems lack the sensitivity necessary to image faint fluorescence.
Flow cytometers also generally require that cells flow through the center of a tightly focused laser beam, which can make them vulnerable to misalignment. Designs that ocean water flows through the instrument. When the scientists retrieve the samples, they inject a dye into them that interacts with the fluorescence and emits colors to reveal their chemical composition.
Medusa has already been deployed several times to study hydrothermal vents, and scientists will continue to send the instrument to the bottom of the ocean in an effort to validate their theory. NASA also plans to test the instrument in other extreme environments of Earth in its continuing quest to seek unknown life forms. Meanwhile, scientists are working to apply the spectral-analysis capabilities of Medusa’s chemical sensor to other areas of research, especially in studying the effects of gravity and cosmic radiation on human cells to possibly create an enhanced system for monitoring the biological effects of space travel on astronauts.
Partnership
To develop the advanced flow cytometry process that is at the crux of Medusa and may one day be part of an advanced astronaut health-monitoring system, NASA sought the help of private industry. The Agency issued a solicitation through the Small Business Innovation Research (SBIR) program at Ames to find a partner for the project. Specifically, NASA needed a partner that could produce a high-speed flow cytometry process to continuously monitor cells for anomalies and bacteria growth. The system would ideally be able to image cells with high-fluorescence sensitivity and would be tolerant of wide variations in sample concentration and other characteristics by having a wide depth of field, allowing cells to be kept in focus regardless of where they happened to be in the flow stream.
Flow cytometry is a powerful technique, but it has several limitations that hinder its use for NASA projects. Most commercial flow cytometers cannot image cells like a microscope. Instead, they measure only the total amount are less sensitive to misalignment tend to sacrifice fluorescence sensitivity.
Many flow cytometry systems are also just too big and impractical for NASA’s purposes, especially since they incorporate pressurized fluid vessels that employ gravity and high pressure to drive the sample through the system.
Amnis Corporation, a Seattle-based biotechnology company, developed a technology called ImageStream for producing sensitive fluorescence images of cells in flow, and happened to be seeking ways to get whole cells into focus in order to increase the usefulness of its systems for research applications. The company had several ideas for how to achieve an extended depth of field, all of which required a level of funding that was just not in its budget. When Amnis heard about the SBIR solicitation, however, the realization came that it could be the perfect opportunity to reap the funding necessary to develop extended depth-of-field technology. The company responded to the SBIR solicitation and proposed to evaluate several methods of extending the depth of field for its ImageStream system, pick the best method, and implement it as an upgrade to its commercial products. This would allow users to view whole cells at the same time, rather than just one section of each cell.
Through Phase I and II SBIR contracts, Ames provided Amnis the funding the company needed to develop this extended functionality. For NASA, the resulting high-speed image flow cytometry process made its way into Medusa and has the potential to benefit space flight health monitoring. On the commercial end, Amnis has implemented the process into its flagship product, ImageStream.
Product Outcome
ImageStream combines high-resolution microscopy and flow cytometry in a single instrument, giving researchers the power to conduct quantitative analyses of individual cells and cell populations at the same time, in the same experiment. Incorporating the extended depth of field developed during the SBIR project, ImageStream is designed to provide multispectral images of rapidly moving objects (in flow) with very high sensitivity, producing up to 6 different microscopic images of each cell flowing through the instrument, at a rate of 15,000 cells per minute. It captures the images at high speeds with a built-in, charge coupled device camera that electronically tracks the motion of cells. A sophisticated auto-focus system continually optimizes image quality.
Getting a whole cell into focus is vital for many different types of cellular analyses, especially for a technique known as fluorescence in situ hybridization (FISH). FISH involves the binding of engineered genetic probes to specific genes or other DNA sequences within cells. The genetic probes consist of a short piece of DNA attached to a fluorescent molecule. The DNA part finds and binds to its complementary DNA sequence(s) within the cell’s genome and the fluorescent molecule signals that the binding has taken place, producing a small spot of light within the cell. By detecting and counting these “FISH” spots, a clinician or a researcher conducting an analysis can tell, for instance, if a patient has extra copies of a gene or a chromosome, whether there is damage to their genes, or whether they have lost genetic material—all of which are very useful for detecting cancer and birth defects.
FISH analysis is primarily a manual process. Clinicians and researchers have to stare through microscopes, adjusting the focus knob until they find all the FISH spots within a given cell. When they do find the spots, they have to manually count them (for about 50 cells per diagnostic test, on average). According to Amnis, this methodology has several problems: the test is slow and expensive, the counting is subjective and prone to error, and, unless a genetic abnormality is present in at least 10 percent of the cells analyzed, researchers will not likely catch it. Automated microscopy systems have been applied to the problem, but they are forced to take multiple images of each cell at different focus settings to ensure that all FISH spots are detected, slowing the test and reducing the economic advantage of automation, according to Amnis.
Prior to the incorporation of the extended depth-of-field technology into the ImageStream system, it was common for one or more FISH spots within a cell to be out of focus as the cell passed through the system. By enhancing the depth of field, Amnis has upgraded ImageStream to accurately detect and count FISH spots through an automated process that eliminates the subjectivity associated with manual counting and can be performed on thousands of cells per minute. This helps drive down the cost of testing by eliminating manual labor, reducing the time needed to perform the test, and increasing accuracy to eliminate retests and false results. Amnis recently announced the availability of this new, high-throughput process.
ImageStream is also built for many other applications, including cell signaling and pathway analysis; classification and characterization of peripheral blood mononuclear cell populations; quantitative morphology; apoptosis (cell death) assays; gene expression analysis; analysis of cell conjugates; molecular distribution; and receptor mapping and distribution. These applications are practical for advanced research in the fields of hematology, immunology, and oncology.
As an add-on option, ImageStream users can pair the instrument with Amnis’ statistical image-analysis software package, IDEAS. This statistical analysis tool is extremely robust, as it provides more than 200 features for every cell analyzed. These features can be used by clinicians and researchers to generate histograms and scatter plots for graphical identification and representation of cells and cell populations based on characteristics like fluorescence intensity, size, shape, texture, probe distribution heterogeneity, and co-localization of multiple probes.
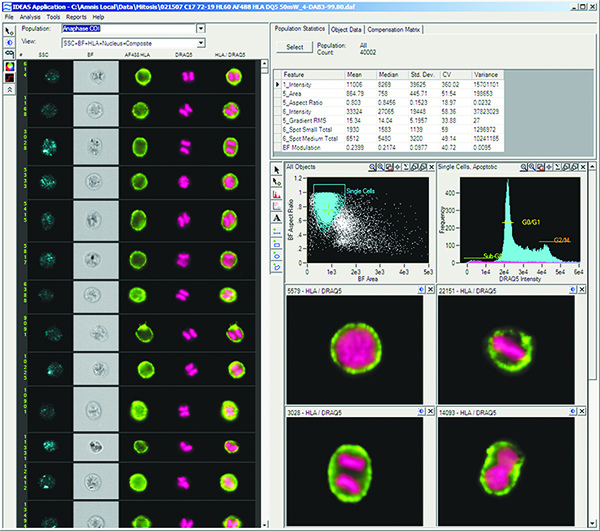
Screen shot of Amnis Corporation’s IDEAS data analysis software showing the detection of cells in division.
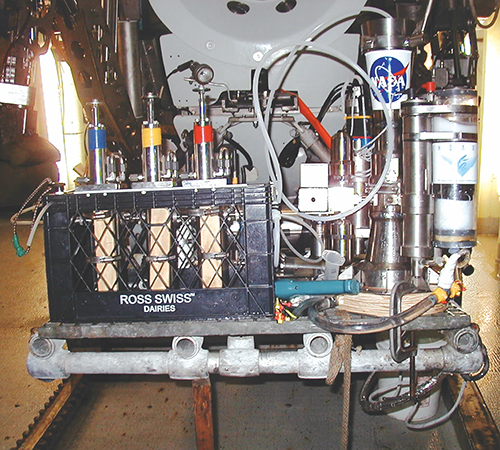
NASA’s Medusa instrument inside the submergence vehicle Alvin’s instrumentation basket in preparation for launch.

The submergence vehicle Alvin as it is about to dive to the bottom of the ocean. Released from the mothership, the Research Vessel Atlantis (operated by Woods Hole Oceanographic Institution, of Woods Hole, Massachusetts) with the diver standing on top.
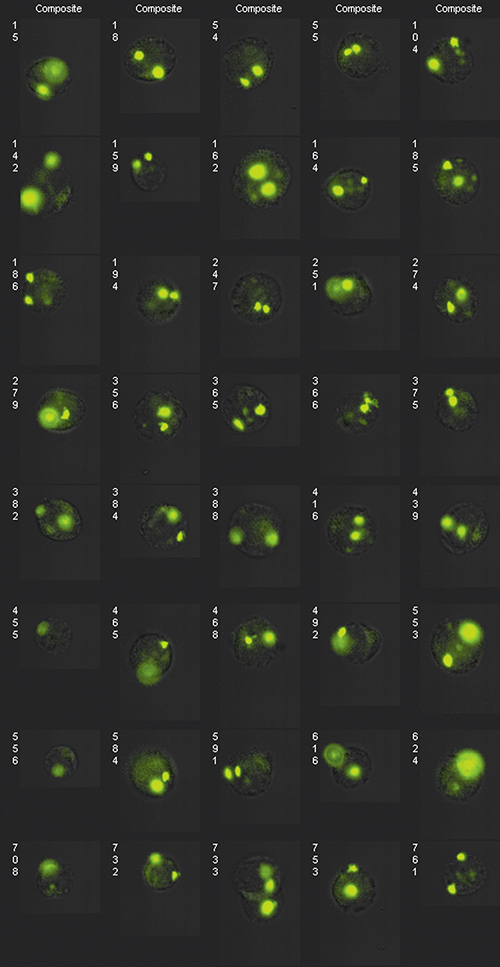
Extended depth-of-field (EDF) image of FISH probes dispersed in the nucleus along the optic axis or in close proximity in the lateral direction. EDF improves image presentation by removing focus-induced variation.

It is possible that beneath Europa’s surface ice, there is a layer of water, kept liquid by tidally generated heat. If so, it would be the only place in the solar system besides Earth where liquid water exists in significant quantities.