Tool Kit Simplifies Development of High-Affinity Molecules
NASA Technology
As NASA sets its sights on long-duration missions deeper into the solar system, including a planned mission to Mars, astronauts will need access to health-monitoring technology. Of particular importance is the need for a diagnostic tool that could measure for biomarkers that indicate loss of bone density, or demineralization, which is a serious concern for astronauts, as living in a microgravity environment causes attrition in the bones and muscles.
But using antibodies, which are proteins commonly utilized to detect biomarkers in diagnostic tests, is problematic, says Dr. Thomas J. Goodwin, who manages the Disease Modeling and Tissue Analogues Laboratory and is lead scientist for Oxidative Stress and Damage research at Johnson Space Center. For one, antibodies in space are susceptible to degradation from radiation exposure. In any case, when refrigerated, antibodies remain viable for only three to six months—far short of the years required for deep-space missions. “Even if you did freeze them, after a couple of years you would have to question their viability,” he notes. “Add to that the fact that you would have to build special storage units into a vehicle that has a lot of space constraints, and it becomes clear that we shouldn’t use antibodies.”
As a result, through the Small Business Innovation Research (SBIR) program, Johnson put out a call for a health-monitoring technology that could not only remain viable for several years, but also be produced simply. One company would contribute to that goal by improving on an alternative to antibodies that has more recently come onto the biotech scene.
Technology Transfer
In the 1990s, researchers discovered through a process called systematic evolution of ligands by exponential enrichment, or SELEX, that short, single strands of RNA and DNA called oligonucleotides could fold themselves into three-dimensional structures that then bind to specific molecules. Called aptamers, they were found to have many potential advantages over antibodies for diagnostics. They can be stored at ambient temperatures without degrading, are impervious to radiation, and, unlike certain antibodies, they don’t provoke a negative immune response in the body, which in extreme cases can be life-threatening.
However, the downside was that, at their stage of development, aptamers did not function as precisely as antibodies in tests, says Mark Shumbera, president of AM Biotechnologies LLC, based in Houston. “They didn’t bind well enough—they weren’t specific enough for their targets. Certain chemical modifications needed to be added to their DNA to make them work better.”
The SELEX process, explains Shumbera, starts with an enormous library of 1014 random oligonucleotide sequences in solution, into which the target molecule is placed. While many sequences will not bind at all to the target, some may bind very well and others will bind weakly. Those sequences that bind are partitioned off and amplified, or copied many times over through a technology called polymerase chain reaction (PCR) in order to create another, smaller library of sequences. After 10 to 15 rounds of partitioning the sequences and amplifying them, only those sequences with the highest binding affinities will remain, and they are identified. While the enzymes used for PCR normally produce unmodified DNA sequences, it is possible to construct sequences that are chemically modified if one or more of the four nucleotides used during PCR contain chemical modifications. Those modifications may improve a sequence’s binding affinity as well as other characteristics.
The problem with this process, says Shumbera, is that the number of chemical modifications possible using the PCR method is limited. Only those modifications the amplifying enzyme will accept can be used, and only a total of four different modifications are possible, since changes made to any nucleobase (adenine, cytosine, guanine, or thymine) are then made to every other base of its kind in the sequence. “But in reality, it’s difficult to use that many,” he says. “The PCR process doesn’t work very efficiently like that, so typically people only use one, and maybe two modifications at a time.”
Through NASA SBIR funding in 2007, AM Biotechnologies advanced what Shumbera describes as a bead-based method for developing aptamers that removes limitations associated with inserting chemical modifications during PCR while also simplifying the process. The technique had been conceptualized several years earlier by David Gorenstein, a chemist and now Associate Dean for Research and Distinguished Chair for Molecular Medicine at the University of Texas Health Science Center in Houston as well as chief science officer for AM Biotechnologies. Through the NASA funding, along with additional support from the Bill & Melinda Gates Foundation and the National Institutes of Health, the company was able not only to create next-generation aptamers with numerous chemical modifications, so-called X-Aptamers, but also to commercialize a kit so others could develop them on their own.
The process begins by utilizing a proprietary technology to synthesize a library of 1010 oligonucleotide sequences, both natural and heavily modified, on microbeads. “You can have 50 modifications in a sequence—there’s virtually no limit,” Shumbera says, compared to the limited capability of the PCR method. “This method allows for the DNA or RNA to be more chemically diverse, meaning there’s a better chance of creating a molecule with a particularly high affinity and specificity for the target.”
Each microbead has billions of copies of just one oligonucleotide sequence synthesized on it, and there is an enormous library of these beads. They’re exposed to the desired target molecule in a solution, and the most promising sequences bind a large number of target molecules to their bead. Magnetic particles that attach to a label on the target molecule are then added to the solution so the beads containing a large amount of the target can be separated from the rest with a magnet. The sequences from those selected beads are cleaved, placed into a solution, and then exposed once again to the target. Shumbera points out this second step—culling the best binders from the solution—only needs to be carried out once because the sheer number of identical sequences on each bead makes finding the highest-affinity sequence easy. “Instead of taking weeks to find the right aptamer with SELEX, our method takes only a few days.”
The sequences of the oligonucleotides pulled out of the solution by the protein target are identified using a next-generation DNA sequencer. “The DNA sequence acts as a barcode to go back to the original library, figure out where the chemical modifications are, and then we resynthesize it. That’s the X-Aptamer,” Shumbera says.
Benefits
After years of further research and development, AM Biotechnologies’ X-Aptamer Selection Kit is now commercially available for any customer, including biotech companies and universities. Practically anyone with basic biochemistry lab skills can work with the technology. “We have university customers, our prototype users, who have had freshman undergraduates select X-Aptamers using our kit. The bead-based process simplifies aptamer selection tremendously,” Shumbera notes.
Beyond their use as a medical diagnostic tool for disease, X-Aptamers can substitute for antibodies as targeting agents for drugs that treat everything from arthritis to cancer. “The X-Aptamer could carry and attach a chemotherapy drug to a tumor, which prevents other parts of the body from being impacted negatively by it,” Shumbera explains, while noting that aptamers themselves can also be made into drugs for treating disease. One such aptamer is Pegaptanib, which is used to treat macular degeneration by preventing the formation of new blood vessels that would otherwise damage the retina.
As far as Shumbera knows, Pegaptanib is the only FDA-approved aptamer drug, and there don’t appear to be many diagnostic tools based on aptamers currently on the market, either. He chalks that up to aptamers being a relatively new technology and their performance not being up to par until now. “The rule of thumb is that it usually takes 20 years for a new technology to really gain mainstream acceptance. It’s been about that long for aptamers. In addition, the chemical modifications that are now being used in our X-Aptamers make them much more effective.”
It’s just the right time, Shumbera says, for something like the X-Aptamer Selection Kit to make its debut. “It has the promise of greatly simplifying the development of affinity-based molecules while also lowering their development cost. It could help usher in the next big revolution in terms of how we diagnose and treat patients.”
For NASA, the X-Aptamer is one critical component for a space-worthy diagnostic tool. The Agency is working with other companies on the other component: a hardware platform that can perform the analysis in space. “No matter what platform or platforms we end up using, right now I think this technology is probably the best solution for the reagent side of the formula,” says NASA’s Goodwin. “X-Aptamers can step up and fill that niche when we’re ready to use it.
Unlike the SELEX method, AM Biotechnologies’ X-Aptamer Selection Kit is simple enough for freshman undergraduates to use and takes only a few days to achieve results.
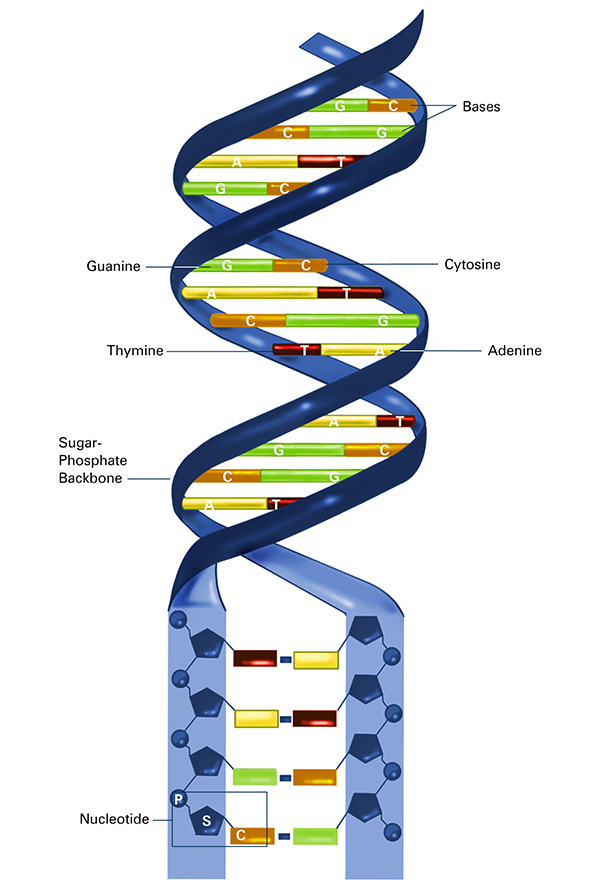
DNA comprises two twisted chains of nucleotides, with each nucleotide containing one base (adenine, thymine, cytosine, or guanine), one phosphate molecule, and the sugar molecule deoxyribose. It was discovered that oligonucleotides—comprising multiple nucleotides—were able to fold themselves into three-dimensional structures called aptamers that, based on their makeup, bind to specific molecules. AM Biotechnologies developed a faster, simplified method for developing highly effective aptamers. Credit: National Institute of General Medical Sciences

An artist’s concept of a human mission to Mars. The area depicted is Noctis Labyrinthus, in the Valles Marineris system of canyons. AM Biotechnologies’ X-Aptamers may be used as part of a health diagnostic tool for missions such as this, which can last for years. On Earth, X-Aptamers can also be used in the development of medicines that aim to treat a range of diseases, from arthritis to cancer. “Instead of taking weeks to find the right aptamer with SELEX, our method takes only a few days.” — Mark Shumbera, AM Biotechnologies